Single-cell systems biology of cancer
Our lab develops experimental and computational methods to unravel regulatory systems on the single-cell level that underlie cancer development. Tumors are a tissue disease: A tumor is an assembly of a myriad of cancer cell clones and their differentiation continua and also includes different types of normal cells. The regulatory circuits of each of these cells are coupled to inter-cellular regulatory systems that rely on cell-to-cell communication and spatial features. The intricate interplay of these coupled control systems has a strong influence on tumor development and ultimately disease progression. Our group’s goal is to develop methods to quantitatively analyze and model trans-cellular circuits to unravel how complex cell phenotypes in tumors are controlled. Our hope is that this will enable targeted modulation that interferes with the hallmarks of cancer and tumor development. You can learn about our previous work on:
Nature webcasts or Genome Medicine and BMC Medicine webinar
We focus on the following research areas:
Novel approaches for single-cell proteomics. To perform systems biology analysis on the single cell level, novel methods for the highly multiplexed analysis of cell phenotypes and signaling networks states are needed. Our group has pioneered a single-cell mass spectrometric approach called mass cytometry (CyTOFTM). This technology allows measurement of levels of over 100 molecules, including proteins and their modifications, on the single-cell level simultaneously with high throughput. The quantitative and correlative nature of mass cytometry data makes this technique ideal for bioinformatics approaches allowing us to identify cell phenotypes and model regulatory circuits. Given the importance of spatial resolution to studies of cellular interplay, we have recently established imaging mass cytometry [you find more information here]. With this approach, we can image dozens of cellular components, including signaling components, simultaneously with subcellular resolution in tissues. Implementing this approach in our current projects is a major effort in the group.
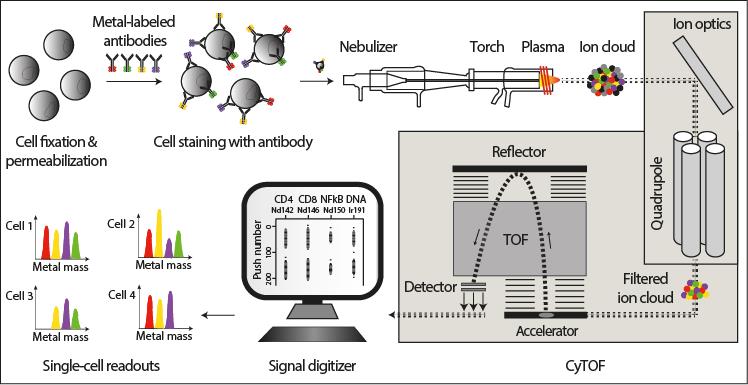
Schematic representation of the mass cytometry workflow (from Di Palma et al. Curr Opin Biotechnol. 2014 Aug 11). Cells of interest are cross-linked by formaldehyde, permeabilized using methanol or detergents, and labeled with antibodies conjugated to pure metal isotopes of defined mass. Subsequently, cells are nebulized, ionized and atomized in a ∼7000 K hot plasma and introduced into the mass cytometer, which is a hybrid quadrupole-time-of-flight mass spectrometer. Here, the composition and quantity of the reporter (metal) isotopes are determined. Signals corresponding to each isotope are then correlated with presence of the respective marker per single-cell event.
Studies of tumor heterogeneity and the tumor microenvironment. To define the coupled cellular and trans-cellular control systems in tumors, the cell types and their spatial assembly must be defined. With the latest single-cell analysis technologies such as mass cytometry, this has now become possible. We combine mass cytometry with ‘omics techniques to systematically describe cell phenotypes and their heterogeneity in tumors. To determine signaling network wiring, we use screening approaches coupled to mass cytometry. Building on these data, imaging mass cytometry is then used to define the spatial interplay of particular cells and their regulatory circuits. We are especially interested in how the processes in the tumor microenvironment induce and maintain the epithelial-mesenchymal transition (EMT), an example of epithelial cell plasticity that may drive cancer metastasis and the generation of cancer stem cells.
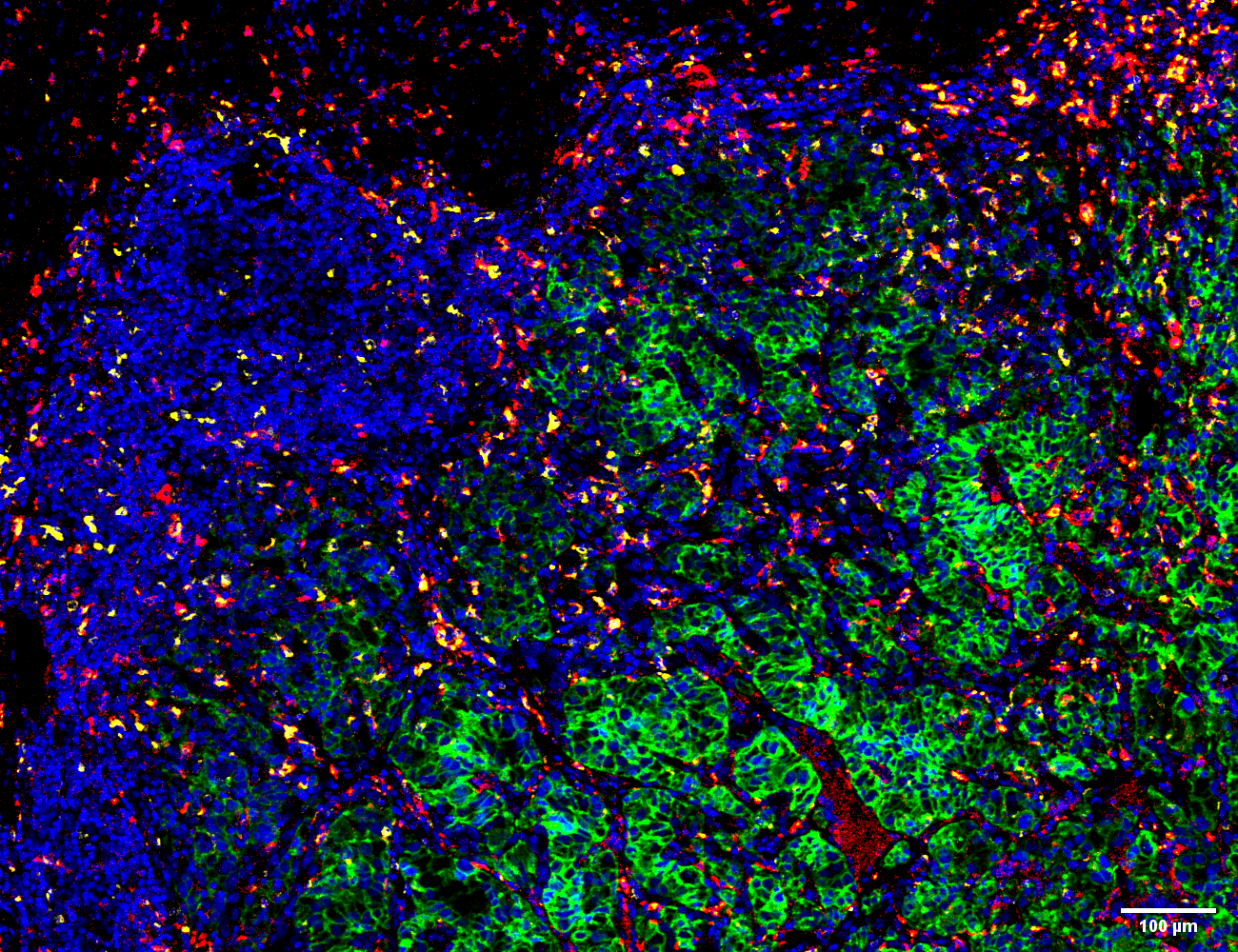
Example of imaging mass cytometry tissue image. In total, 40 proteins and phosphorylation sites were measured simultaneously at 1-μm resolution. Visualized pseudo-color overlay of carbonic anhydrase (green), CD206 (red), CD68 (yellow) and DNA (blue).
Modeling of regulatory circuits in cancer. The single-cell systems biology analyses of patient tumor samples yields a wealth of data about cancer. In addition to developing computational tools to process the generated data, we use a variety of algorithms and data driven modeling approaches to model subpopulation of cells and their signaling network structures. Further, using data from imaging mass cytometry, we model how signaling network states couple with those of other cells in a spatially resolved manner. In a systems biology manner, we then use these models to predict how to experimentally refine the computational models until convergence is reached. By modeling of the regulatory circuits of cancer stem cells generated via EMT, we have identified key regulatory elements that are responsible for the control of the transition. Modeling of the imaging mass cytometry data revealed how the hypoxic state in the tumor core differentially affects tumor subclones.
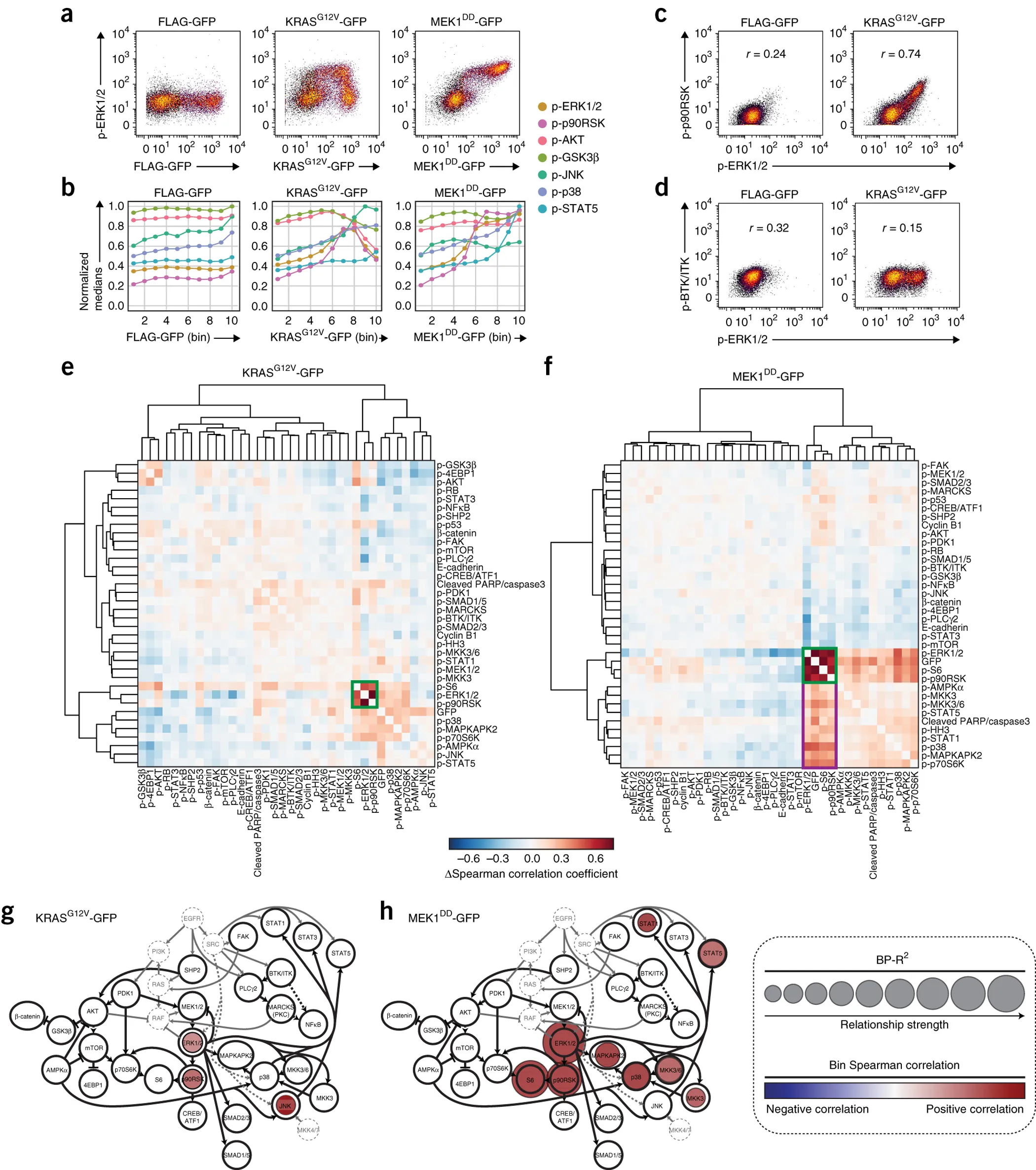
MAPK/ERK pathway mutants induce oncogenic signaling. (Figure from Lun XK et al., Nature Biotechnology, 35, pages164–172(2017)).